The tidyverse (opens in new tab) is one of my favourite suites of R packages, and its idiomatic approach to data manipulation, analysis and visualization has heavily influenced how I approach data problems in R. If there’s one tidyverse package that best embodies these qualities, my vote is for purrr (opens in new tab)’s clean approach to iteration. purrr goes well beyond map and its variants (check out this presentation for a great sampler (opens in new tab)), but it’s been far too easy to ignore anything beyond purrr’s simplest functions.
A while ago at work I had a great opportunity to dive deeper into purrr. While demonstrating the Central Limit Theorem to some
colleagues, I whipped up a simulation of the dice-rolling experiment in R. I didn’t save the code, but it looked a lot like this:
library(tidyverse)library(purrr)library(rlang)library(daniel) # see https://gitlab.com/danielspracklin/daniel
simulate_dice <- function(n, number_of_dice, ...) { seq.int(number_of_dice) %>% map(~sample(1:6, n, replace = T)) %>% as.data.frame(row.names = NULL) %>% rowwise() %>% summarize(average_of_dice = sum(c_across(where(is.numeric))) / {{number_of_dice}}, number_of_dice = {{number_of_dice}})}
clt_plotter <- function(df, x_value, facet_value) { df %>% ggplot() + geom_density(aes({{x_value}}, fill = "1"), colour = "black") + scale_fill_daniel() + facet_wrap(vars({{facet_value}})) + theme_daniel() + guides(fill = F) + labs(x = "Sampling distribution of the mean", y = "Density")}And here’s the output. The wiggles in the number_of_dice = 1 facet are a necessary evil if we want to show the smooth behaviour for larger numbers of dice without customizing the density plots’ bandwidth and kernel.
set.seed(613)1:9 %>% map_dfr(~simulate_dice(10000, .x)) %>% clt_plotter(., x_value = average_of_dice, facet_value = number_of_dice)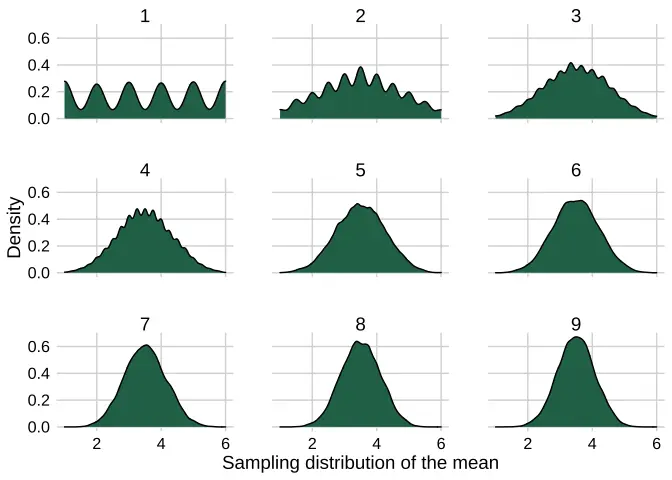
The simulate_dice code is functional but hacky:
- Heavy reliance on dataframes here is a crutch, since we have to use the very slow
rowwiseoperation to create a column containing the mean of each set of thrown dice. - Inserting the number of dice thrown as its own column is an ugly way to prepare to facet the
clt_plottervisualization.
We can do better than this, and in the process learn more about the power of purrr.
We’ll start by writing a function that doesn’t immediately coerce everything to a dataframe. Instead, we’ll keep the results as a list as long as possible. To add the vectors element-wise, we can use purrr::reduce to avoid rowwise and mutate. (Since addition is associative, we don’t need to specify the .dir argument for reduce.) I also wrote a division function, vector_division, to get the mean of each set of thrown dice, although there’s likely a more idiomatic way to do this.
simulate_dice_list <- function(n, number_of_dice, ...) {
vector_division <- function(x, divisor) {x / divisor}
seq.int(number_of_dice) %>% map(~sample(1:6, n, replace = T)) %>% reduce(`+`) %>% map(~vector_division(.x, {{number_of_dice}})) %>% unlist()}Now let’s compare the outputs to confirm that they’re identical. Since the new version doesn’t produce a dataframe, we’ll have to wrangle it into shape. enframe gives us a nested dataframe that requires unnest-ing. As we can see, the results are identical.
set.seed(613)old_version <- 1:9 %>% map_dfr(~simulate_dice(10000, .x))
set.seed(613)new_version <- 1:9 %>% map(~simulate_dice_list(10000, .x)) %>% enframe(.) %>% unnest(value)
identical(old_version$average_of_dice, new_version$value)
## [1] TRUESo which is better? It all comes down to performance, which I assessed quickly with microbenchmark.
library(microbenchmark)
set.seed(613)benchmark <- microbenchmark( old_version = 1:9 %>% simulate_dice(100, .x), new_version = 1:9 %>% simulate_dice_list(100, .x) %>% enframe(.) %>% unnest(value))
benchmark
## Unit: milliseconds## expr min lq mean median uq max neval## old_version 23.097471 36.879219 57.42355 54.53358 72.07341 131.73285 100## new_version 5.350244 7.995392 15.33288 13.58652 19.48240 67.29409 100
autoplot(benchmark) + theme_daniel()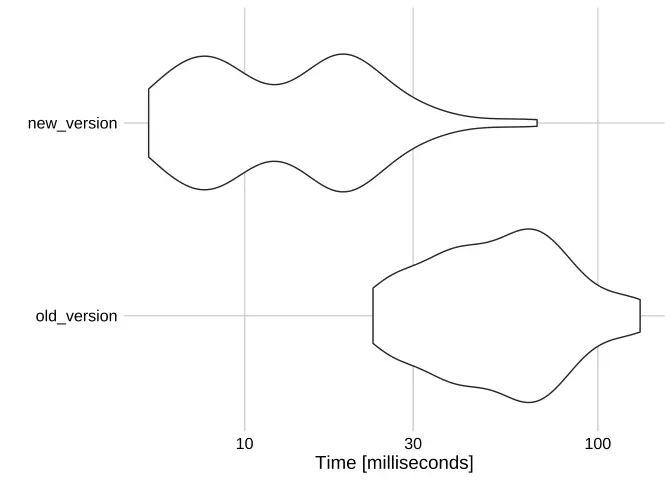
The difference is drastic: rowwise, while easy to pluck from the grab bag of dplyr tools, is indeed much slower than using lists.
Here’s the final code. As a bonus, we can avoid the summarize(number_of_dice = {{number_of_dice}})
code above by naming the input vector with setNames.
set.seed(613)setNames(c(1:9), c(1:9)) %>% map(~simulate_dice_list(10000, .x)) %>% enframe(.) %>% unnest(value) %>% clt_plotter(., x_value = value, facet_value = name)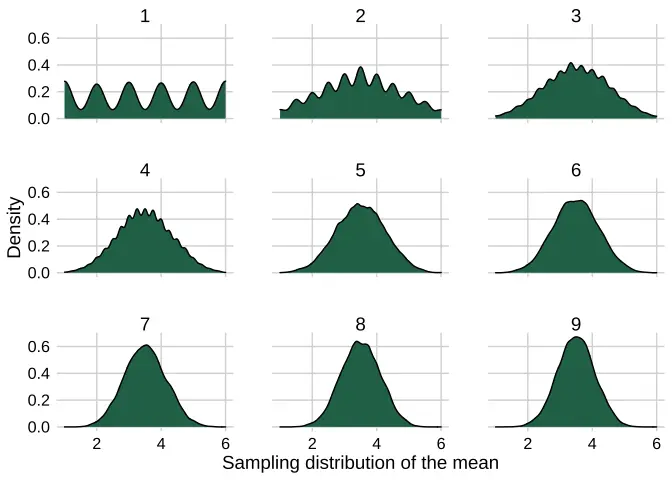
No difference in output, but faster and cleaner. It pays to use purrr in conjunction with more than just dataframes!